publications
research articles and datasets
2025
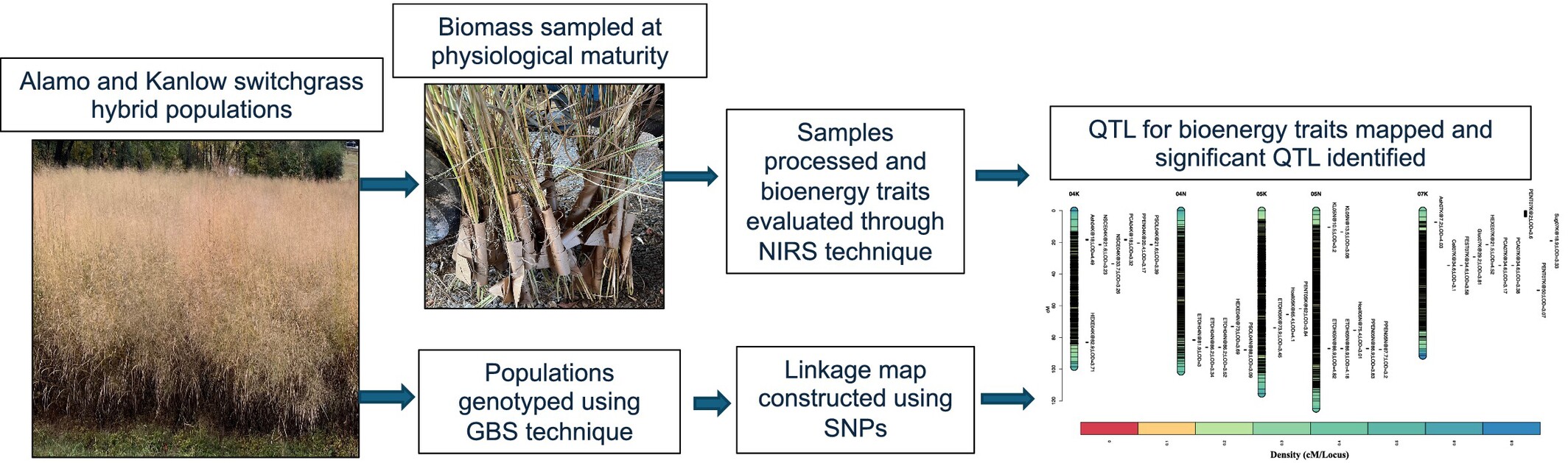
- Mapping Quantitative Trait Loci for Bioenergy Traits in Multiple Hybrid Populations of Lowland Switchgrass in Simulated-Sward PlantingSurya L. Shrestha, Christian M. Tobias, Fred Allen, Jennifer Bragg, Ken Goddard, and Hem S. BhandariGlobal Change Biology Bioenergy, 2025e70060 GCB-B-RA-24-154.R1
Switchgrass (Panicum virgatum L.) is a potential source of producing bioenergy from lignocellulosic biomass. Bioenergy traits are quantitatively inherited. This study localized variation in bioenergy traits estimated via near infrared spectroscopy using quantitative trait loci (QTL) mapping. Eight hybrid populations (30 to 96 F1s) developed by crossing lowland cultivars, Alamo and Kanlow, were evaluated in two environments in Tennessee using a randomized complete block design with two replications per location in 2020 and 2021. The hybrid populations exhibited significant variation for all the studied traits (p ≤ 0.05). A linkage map including 17,251 single nucleotide polymorphisms (SNPs) generated through genotype-by-sequencing was used for the QTL mapping. The QTL analyses were performed on the traits across populations in each and across environments (years and locations) and detected a total of 74 significant QTL peaks with the logarithm of odds (LOD) scores ranging from 3.0 to 6.9. Phenotypic variability explained (PVE) by QTL varied from 2.1% to 7.4%. Ten QTL for predicted ethanol yield were identified on chromosomes 4N, 5K, 5N, 8K, 8N, and 9N, respectively, in which the major QTL resided on chromosome 5N with the highest PVE value (7.4%). Four cellulose and three hemicellulose QTL were identified on chromosomes 1K, 1N, 2N, 5K, 5N, 7K, and 8N, with PVE ranging from 2.1% to 5.8%. The chromosomal regions of 1N, 4K, 5N, and 7K had pleiotropic effects affecting multiple bioenergy traits. SNPs linked to QTL will be useful for improving bioenergy traits through marker-assisted breeding.
@article{shrestha_gcb_2025, author = {Shrestha, Surya L. and Tobias, Christian M. and Allen, Fred and Bragg, Jennifer and Goddard, Ken and Bhandari, Hem S.}, title = {Mapping Quantitative Trait Loci for Bioenergy Traits in Multiple Hybrid Populations of Lowland Switchgrass in Simulated-Sward Planting}, journal = {Global Change Biology Bioenergy}, volume = {17}, number = {9}, pages = {e70060}, keywords = {bioenergy, genotype by environment interaction, genotype by sequencing, hybrid, Panicum virgatum, quantitative trait loci, single nucleotide polymorphism}, doi = {10.1111/gcbb.70060}, url = {https://onlinelibrary.wiley.com/doi/abs/10.1111/gcbb.70060}, eprint = {https://onlinelibrary.wiley.com/doi/pdf/10.1111/gcbb.70060}, note = {e70060 GCB-B-RA-24-154.R1}, urldate = {2025-08-11}, dimensions = {true}, year = {2025}, project = switchgrass } - Bioenergy traits data of lowland switchgrass (Panicum virgatum L.) hybrid populationsSurya Shrestha, Christian Tobias, Fred Allen, Jennifer Bragg, Ken Goddard, and Hem BhandariJun 2025
Switchgrass (Panicum virgatum L.) is a potential source for producing bioenergy from lignocellulosic biomass. Many breeding programs focus on the genetic improvement of switchgrass for increasing bioenergy traits. Significant genetic variation for bioenergy traits was observed in lowland switchgrass hybrids (P<0.05). Due to the quantitative inheritance of bioenergy traits, varietal improvement for the trait through conventional methods is slow and challenging. Therefore, quantitative trait loci (QTL) mapping is used to discover marker-trait associations and accelerate the breeding process through marker-assisted selection. To identify significant QTL, this study mapped eight hybrid populations (30 to 96 F1s) developed by crossing lowland cultivars, Alamo and Kanlow. The populations were evaluated in a simulated-sward plot with two replications at two locations in Tennessee in 2020 and 2021. The crosses were genotyped using 17,251 single nucleotide polymorphisms generated through genotyping-by-sequencing. QTL mapping was performed across populations using R-QTL. The study identified ten QTL for predicted ethanol, four for cellulose, and three hemicellulose on chromosomes 1K, 1N, 2N, 4N, 5K, 5N, 7K, 8K, 8N, and 9N, respectively, with phenotypic variability ranged from 2.1 to 7.4%. The dataset contains two comma-separated files (CSV) describing the phenotype of each individual used in the quantitative trait loci (QTL) analysis. The genotype data, including the number of SNPs and thier chromosomal locations, progeny file and linkage map, are provided in Shrestha et al. (2023).
@misc{shrestha_bioenergy_2025, title = {Bioenergy traits data of lowland switchgrass ({Panicum} virgatum {L}.) hybrid populations}, copyright = {Creative Commons Attribution 4.0 International}, url = {https://zenodo.org/doi/10.5281/zenodo.15770211}, doi = {10.5281/ZENODO.15770211}, urldate = {2025-07-25}, publisher = {Zenodo}, datacite = {true}, author = {Shrestha, Surya and Tobias, Christian and Allen, Fred and Bragg, Jennifer and Goddard, Ken and Bhandari, Hem}, month = jun, year = {2025} }
2023
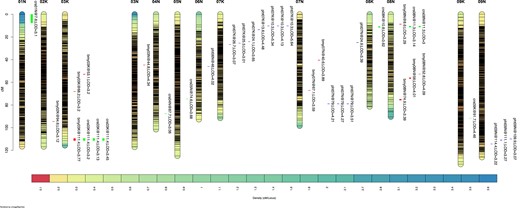
- Mapping quantitative trait loci for biomass yield and yield-related traits in lowland switchgrass (Panicum virgatum L) multiple populationsSurya L. Shrestha, Christian M Tobias, Hem S Bhandari, Jennifer Bragg, Santosh Nayak, Ken Goddard, and Fred AllenG3: Genes, Genomes, Genetics, May 2023
Switchgrass can be used as an alternative source for bioenergy production. Many breeding programs focus on the genetic improvement of switchgrass for increasing biomass yield. Quantitative trait loci (QTL) mapping can help to discover marker-trait associations and accelerate the breeding process through marker-assisted selection. To identify significant QTL, this study mapped 7 hybrid populations and one combined of 2 hybrid populations (30–96 F1s) derived from Alamo and Kanlow genotypes. The populations were evaluated for biomass yield, plant height, and crown size in a simulated-sward plot with 2 replications at 2 locations in Tennessee from 2019 to 2021. The populations showed significant genetic variation for the evaluated traits and exhibited transgressive segregation. The 17,251 single nucleotide polymorphisms (SNPs) generated through genotyping-by-sequencing (GBS) were used to construct a linkage map using a fast algorithm for multiple outbred families. The linkage map spanned 1,941 cM with an average interval of 0.11 cM between SNPs. The QTL analysis was performed on evaluated traits for each and across environments (year and location) that identified 5 QTL for biomass yield (logarithm of the odds, LOD 3.12–4.34), 4 QTL for plant height (LOD 3.01–5.64), and 7 QTL for crown size (LOD 3.0–4.46) (P ≤ 0.05). The major QTL for biomass yield, plant height, and crown size resided on chromosomes 8N, 6N, and 8K explained phenotypic variations of 5.6, 5.1, and 6.6%, respectively. SNPs linked to QTL could be incorporated into marker-assisted breeding to maximize the selection gain in switchgrass breeding.
@article{shrestha_mapping_2023, title = {Mapping quantitative trait loci for biomass yield and yield-related traits in lowland switchgrass ({Panicum virgatum} {L}) multiple populations}, volume = {13}, copyright = {https://creativecommons.org/licenses/by/4.0/}, issn = {2160-1836}, url = {https://academic.oup.com/g3journal/article/doi/10.1093/g3journal/jkad061/7083855}, doi = {10.1093/g3journal/jkad061}, language = {en}, number = {5}, urldate = {2025-07-24}, dimensions = {true}, journal = {G3: Genes, Genomes, Genetics}, author = {Shrestha, Surya L. and Tobias, Christian M and Bhandari, Hem S and Bragg, Jennifer and Nayak, Santosh and Goddard, Ken and Allen, Fred}, editor = {Brown, P}, month = may, year = {2023}, project = switchgrass } - Biomass Bioenergy
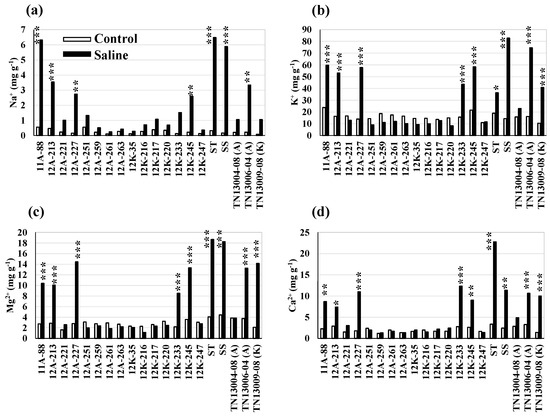
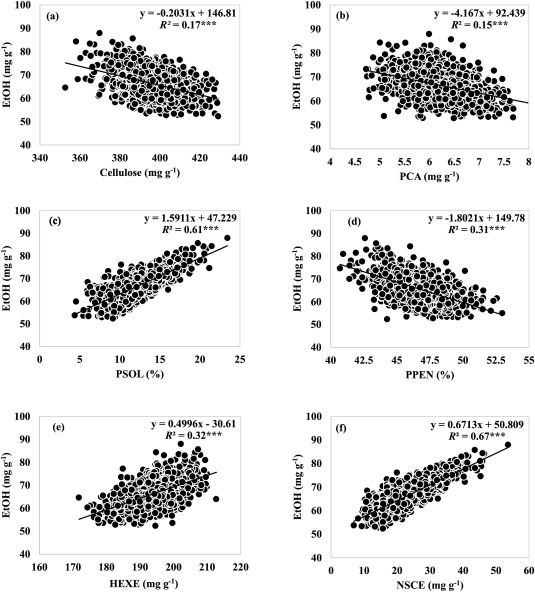 Genetic variation for bioenergy traits within and among lowland switchgrass (Panicum virgatum L.) crossesSurya L. Shrestha, Fred L. Allen, Ken Goddard, Hem S. Bhandari, and Gary E. BatesBiomass and Bioenergy, Aug 2023
Genetic variation for bioenergy traits within and among lowland switchgrass (Panicum virgatum L.) crossesSurya L. Shrestha, Fred L. Allen, Ken Goddard, Hem S. Bhandari, and Gary E. BatesBiomass and Bioenergy, Aug 2023Cellulosic biomass derived from switchgrass can be an alternative source to produce renewable bioenergy. However, bioenergy production from switchgrass is not economically viable unless genotypes with high yield potential and acceptable biofuel quality are available. To identify switchgrass genotypes with good biofuel quality, ten high biomass yielding hybrid populations derived from crosses between two lowland cultivars, Alamo (A) and Kanlow (K), were evaluated in this study. The experiment was planted in 2018 in two field environments, the East Tennessee Research and Education Center, Knoxville, and the Plateau Research and Education Center, Crossville, in a randomized complete block design with two replications per location. Tillers were collected in the Fall of 2020 and 2021, and analyzed for bioenergy traits using the near-infrared reflectance spectroscopy technique. The crosses and their parents were different in bioenergy traits across the environments (locations and years) (P ≤ 0.05). The crosses’ total ethanol yield (EtOH) ranged from 63.5 to 70.4 mg g−1, and the total sugars yield varied from 725 to 737 mg g−1 across the environments. Average cellulose, hemicellulose, lignin, and ash content were 395.8, 333.9, 178.4, and 49.3 mg g−1, respectively. The estimated non-structural carbohydrates were found as good predictors of EtOH (R2 = 0.67) (P ≤ 0.05). The cross, 12A-259 × 12K-247, exhibited high EtOH, high hemicellulose, low lignin, and low ash. Crosses 12A-261 × 12K-245 and 12A-263 × 12K-250 exhibited high cellulose and sugar. These results indicate significant genetic heterogeneity for bioenergy traits within and between these two lowland switchgrass cultivars.
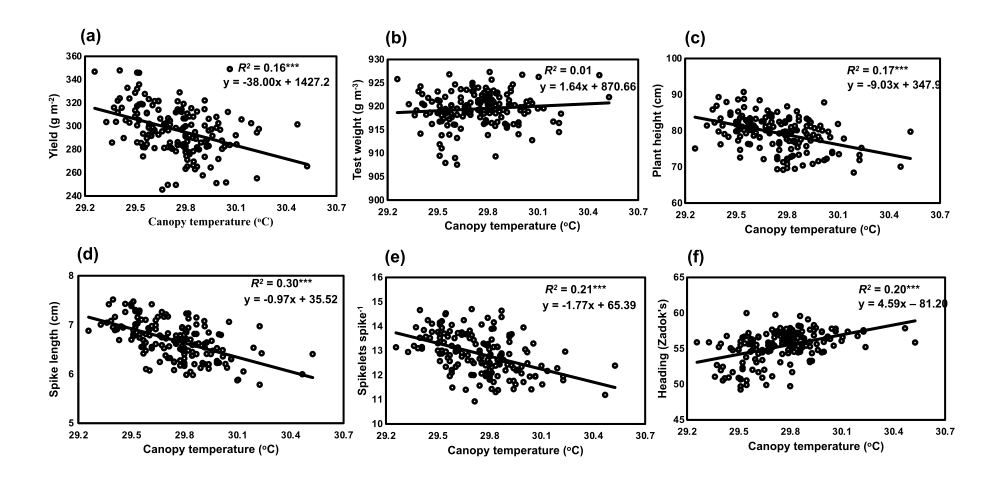
@article{shrestha_genetic_2023, title = {Genetic variation for bioenergy traits within and among lowland switchgrass ({Panicum} virgatum {L}.) crosses}, volume = {175}, issn = {09619534}, url = {https://linkinghub.elsevier.com/retrieve/pii/S0961953423001769}, doi = {10.1016/j.biombioe.2023.106878}, language = {en}, urldate = {2025-07-24}, dimensions = {true}, journal = {Biomass and Bioenergy}, author = {Shrestha, Surya L. and Allen, Fred L. and Goddard, Ken and Bhandari, Hem S. and Bates, Gary E.}, month = aug, year = {2023}, pages = {106878}, project = switchgrass } - Association of canopy temperature with agronomic traits in spring wheat inbred populationsSurya L. Shrestha, Kimberly A. Garland-Campbell, Camille M. Steber, William L. Pan, and Scot H. HulbertEuphytica, Jan 2023
Canopy temperature (CT) is considered a reliable proxy for stomatal conductance. Low CT values of plant canopies under water-limited conditions are associated with high transpiration indicating plants’ drought tolerance. Many U.S. Pacific Northwest (PNW) adapted wheat (Triticum aestivum L.) cultivars lack stress-adaptive traits resulting in poor performance in drought environments. This study aims to identify the stress-adaptive traits by evaluating the CT in spring wheat populations across different soil moisture conditions in the PNW. An infrared thermometer was used to estimate the CT in two families of recombinant inbred lines, ‘Alpowa’ × ‘Express’ (AE population) and ‘Hollis’ × ‘Drysdale’ (HD population), in rainfed and irrigated environments of the dryland PNW in 2011 to 2013. Higher reductions in grain yield up to 170%, spike length up to 25%, and spikelets spike−1 up to 19% were observed in a rainfed environment compared to the reductions in an irrigated environment. A significant variation in CT was observed in both AE and HD populations. With 1 °C increase in CT at the anthesis stage, grain yield was lowered up to 38 g m−2. Low CT was associated with high grain yield and agronomic traits in both wheat populations (r = − 0.18 to − 0.55, P ≤ 0.05). The highest association between CT and grain yield was observed at anthesis (r = − 0.47) and milking (r = − 0.38) stages (P ≤ 0.001). Our results show that screening for low CT during terminal wheat growth stage is an effective strategy for improving the selection of new drought-tolerant wheat varieties in the PNW.
@article{shrestha_association_2023, title = {Association of canopy temperature with agronomic traits in spring wheat inbred populations}, volume = {219}, issn = {0014-2336, 1573-5060}, url = {https://link.springer.com/10.1007/s10681-022-03135-4}, doi = {10.1007/s10681-022-03135-4}, language = {en}, number = {1}, urldate = {2025-07-24}, journal = {Euphytica}, dimensions = {true}, author = {Shrestha, Surya L. and Garland-Campbell, Kimberly A. and Steber, Camille M. and Pan, William L. and Hulbert, Scot H.}, month = jan, year = {2023}, pages = {7}, project = wheat } - Alamo x Kanlow genotypic and phenotypic data for biomass yield and yield-related traits in lowland switchgrass (Panicum virgatum L.) crossesSurya Shrestha, Christian M. Tobias, Hem S. Bhandari, Jennifer Bragg, Santosh Nayak, Ken Goddard, and Fred AllenMar 2023
Switchgrass (Panicum virgatum L.) is a model herbaceous bioenergy crop in the USA. It is a native, perennial, warm-season grass, and has broad adaptability. Many breeding programs focus on the genetic improvement of switchgrass for increasing biomass yield. Significant genetic variation for biomass yield observed in lowland switchgrass hybrids. Due to the quantitative inheritance of biomass yield, varietal improvement for the trait through conventional breeding is slow. Therefore, quantitative trait loci (QTL) mapping is used to discover marker-trait associations and accelerate the breeding process through marker-assisted selection. To identify significant QTL, this study mapped seven biparental crosses and one combined cross of two biparental crosses (30 to 96 F1s) between lowland Alamo and Kanlow genotypes. The crosses were evaluated for biomass yield, plant height, and clonal mass scores in a simulated-sward plot with two replications at two locations in Tennessee from 2019 to 2021. The crosses were genotyped using 17,251 single nucleotide polymorphisms generated through genotyping-by-sequencing. QTL mapping was performed using a single-QTL model in R-QTL. The study identified major QTL for biomass yield, plant height, and clonal mass scores resided on chromosomes 7K, 4K, and 3K and had 0.47, 0.63, and 0.62 heritability, respectively. The dataset contains five files describing the phenotype and genotype of each individual used in the quantitative trait loci (QTL) analysis.
@misc{shrestha_alamo_2023, title = {Alamo x {Kanlow} genotypic and phenotypic data for biomass yield and yield-related traits in lowland switchgrass ({Panicum} virgatum {L}.) crosses}, copyright = {Creative Commons Zero v1.0 Universal}, url = {https://datadryad.org/dataset/doi:10.5061/dryad.hmgqnk9mm}, doi = {10.5061/DRYAD.HMGQNK9MM}, language = {en}, urldate = {2025-07-25}, publisher = {Dryad}, datacite = {true}, author = {Shrestha, Surya and Tobias, Christian M. and Bhandari, Hem S. and Bragg, Jennifer and Nayak, Santosh and Goddard, Ken and Allen, Fred}, month = mar, year = {2023}, keywords = {Biomass, experimental hybrids, FOS: Agricultural sciences, Panicum virgatum L., QTL, switchgrass} }
2022
- Genotypic Variation for Salt Tolerance within and between Alamo and Kanlow Switchgrass (Panicum virgatum L.) CultivarsSurya L. Shrestha, Carl Sams, and Fred AllenAgronomy, Mar 2022
Switchgrass tolerates nutrient and water limitations; however, high salt concentrations may inhibit its production. Therefore, the salt tolerance potential of switchgrass needs to be enhanced for economic production. The objective of this study was to determine the differences within and between two lowland cultivars of switchgrass for growth, stomatal morphology, photosynthetic pigments content, and mineral traits. A greenhouse study was conducted on 18 sublines, derived from the cultivars Alamo (A) and Kanlow (K), plus a salt-tolerant and sensitivity check (20 genotypes). A split-plot randomized complete block design was used with three replications per treatment (0, 5, 10, and 20 dS m−1 NaCl). High salt concentrations (10 and 20 dS m−1 NaCl) reduced shoot biomass, stem diameter, and plant height up to 21, 11, and 16%, respectively, compared to the control for all genotypes (p \textless 0.05). Chlorophyll a and adaxial stomata width were moderately correlated with growth traits under saline conditions (r = 0.49 to 0.56, p \textless 0.05). The sublines 12A-227, TN13006-04, 12A-259, 12K-247, and TN13009-08 had better growth and accumulated less salt in shoot biomass than both checks. These results indicate that differences exist within and between the Alamo and Kanlow sublines, and it is possible to breed improved cultivars with increased salt tolerance.
@article{shrestha_genotypic_2022, title = {Genotypic {Variation} for {Salt} {Tolerance} within and between {Alamo} and {Kanlow} {Switchgrass} ({Panicum} virgatum {L}.) {Cultivars}}, volume = {12}, copyright = {https://creativecommons.org/licenses/by/4.0/}, issn = {2073-4395}, url = {https://www.mdpi.com/2073-4395/12/4/973}, doi = {10.3390/agronomy12040973}, language = {en}, number = {4}, urldate = {2025-07-24}, dimensions = {true}, journal = {Agronomy}, author = {Shrestha, Surya L. and Sams, Carl and Allen, Fred}, year = {2022}, pages = {973}, project = switchgrass }
2021
- Heterosis for biomass yield and other traits in ‘Alamo’ × ‘Kanlow’ switchgrass populationsSurya L. Shrestha, Hem S. Bhandari, Fred L. Allen, Christian M. Tobias, Santosh Nayak, Ken Goddard, and Scott A. SensemanCrop Science, Nov 2021
Heterosis plays an important role in switchgrass (Panicum virgatum L.) breeding. To identify superior clones for hybrid development through seasonal single cut, 10 crosses derived from ‘Alamo’ × ‘Kanlow’ and parents were evaluated. The experiment was planted in 2018 using a randomized complete block design with two replications at two locations in Tennessee: the East Tennessee Research and Education Center (ETREC), Knoxville, and the Plateau Research and Education Center (PREC), Crossville. The crosses and their parents were evaluated for biomass yield, plant height, and clonal mass in the fall of 2019 and 2020. The crosses were significantly different in biomass yield within and across locations (P ≤ .05). Average biomass yield of the crosses ranged from 7.1 to 8.7 Mg ha−1 at ETREC and from 10.0 to 11.8 Mg ha−1 at PREC. The crosses demonstrated an average of 46% mid-parent heterosis (MPH) and 24% high-parent heterosis (HPH) for biomass yield across the two locations. The MPH and HPH were 3 and −1.3% for plant height and 12 and 1% for clonal mass, respectively. Moderate to strong positive association was found between clonal mass scores and biomass yield at individual locations across years and across locations and years (r = 0.37–0.79, P ≤ .05). The association between biomass yield and plant height was inconsistent (r = −0.24 to 0.67, P ≤ .05). The findings of biomass yield heterosis in these crosses suggested the potential of hybrid cultivars for biomass yield improvement in lowland switchgrass.
@article{shrestha_heterosis_2021, title = {Heterosis for biomass yield and other traits in ‘{Alamo}’ × ‘{Kanlow}’ switchgrass populations}, volume = {61}, issn = {0011-183X, 1435-0653}, url = {https://acsess.onlinelibrary.wiley.com/doi/10.1002/csc2.20618}, doi = {10.1002/csc2.20618}, language = {en}, number = {6}, urldate = {2025-07-24}, journal = {Crop Science}, dimensions = {true}, author = {Shrestha, Surya L. and Bhandari, Hem S. and Allen, Fred L. and Tobias, Christian M. and Nayak, Santosh and Goddard, Ken and Senseman, Scott A.}, month = nov, year = {2021}, pages = {4066--4080}, project = switchgrass }
2020
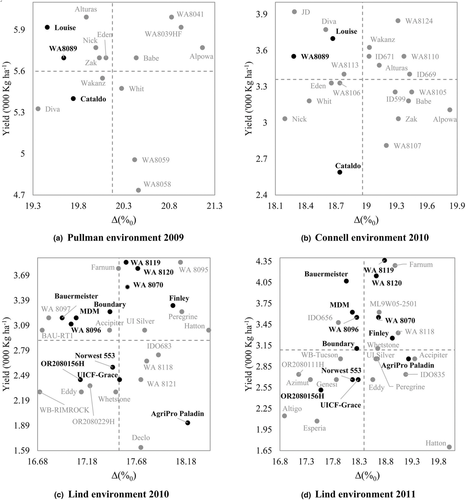
- Carbon isotope discrimination association with yield and test weight in Pacific Northwest–adapted spring and winter wheatSurya L. Shrestha, Kimberly A. Garland‐Campbell, Camille M. Steber, and Scot H. HulbertAgrosystems, Geosciences & Environment, Jan 2020
Due to considerable influence of environment on yield, breeding for drought tolerance could benefit from focusing on selection of more heritable physiological traits, such as carbon isotope discrimination (as measured by delta, ∆) for indirectly assessing water use efficiency (WUE) of wheat (Triticum aestivum L.). Spring and winter wheat cultivars were assayed for ∆, and these values were used to determine the relationships with performance in over 13 environments in the U.S. Pacific Northwest. The correlation coefficients of ∆ values between the wheat cultivars grown in different environments ranged from 0.11 to 0.73 for both spring and winter wheat. There was significant genotypic variation for ∆ in soft spring and hard winter wheat but not in hard spring and soft winter wheat. The ∆ values were poor indicators of yield for this set of wheat cultivars in most environments, although low values (better WUE) were sometimes correlated with yield. A population of 165 hard spring wheat recombinant inbred lines derived from a cross between two hard spring wheat varieties that differed in ∆ was also screened in low-rainfall dryland and irrigated environments. High yields in this recombinant inbred population were weakly correlated with high ∆ values or low WUE in most environments.
@article{shrestha_carbon_2020, title = {Carbon isotope discrimination association with yield and test weight in {Pacific} {Northwest}–adapted spring and winter wheat}, volume = {3}, issn = {2639-6696, 2639-6696}, url = {https://acsess.onlinelibrary.wiley.com/doi/10.1002/agg2.20052}, doi = {10.1002/agg2.20052}, language = {en}, number = {1}, urldate = {2025-07-24}, dimensions = {true}, journal = {Agrosystems, Geosciences \& Environment}, author = {Shrestha, Surya L. and Garland‐Campbell, Kimberly A. and Steber, Camille M. and Hulbert, Scot H.}, month = jan, year = {2020}, pages = {e20052}, project = wheat }